Table of Contents

Purpose
We develop MiiL (Medical Image Illustrator) to help users see the unseen, and provide many image processing and visualization functionality. Users can play with these tools similar to the interaction style of the popular image editing software and explore the data.
Abstract
MiiL is a medical image visualization system for diagnosis and surgical planning. This software reads the DICOM images directly and perform semi-automatic segmentation, measurement and other image adjustment processes. In the post-processing stage, it allows users to visualize the volume data with transfer function adjustment to highlight the image feature. MiiL lets the medical staffs interactively manipulate the data and provide visual effect that helps the communication between the patients and doctors.

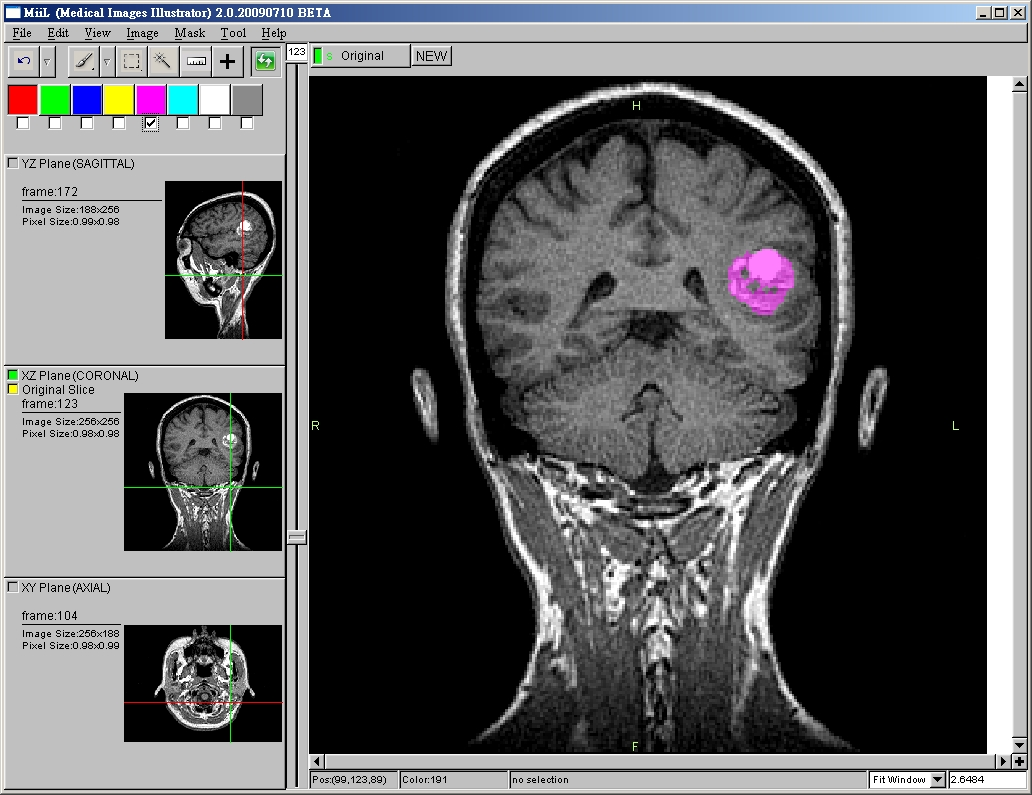

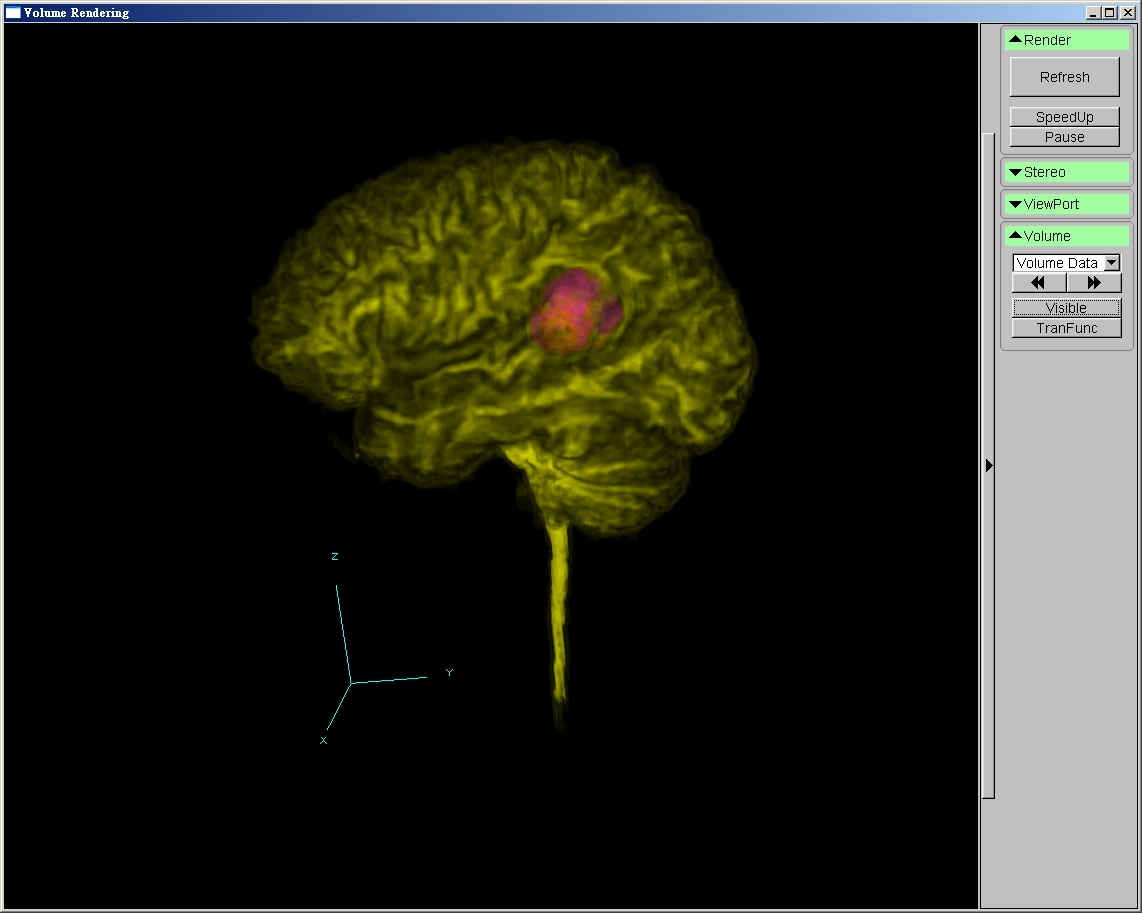
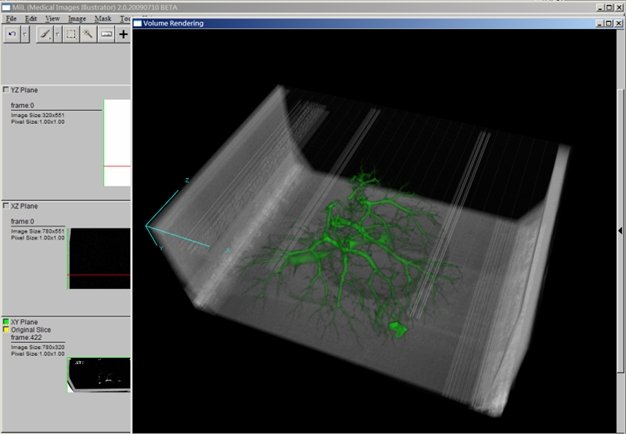
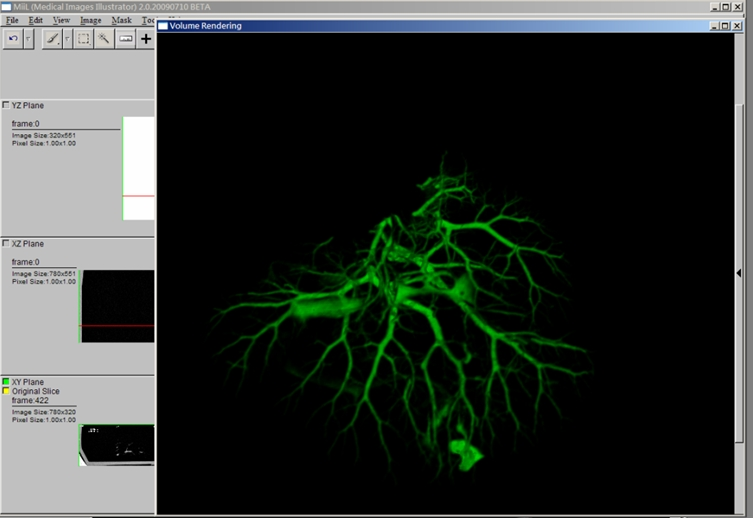
Goals
We want to develop a medical image visualization system that allows the medical staffs to use the software on PC for diagnosis, training, surgical planning and even research. This system must have some friendly and familiar user interface without sacrificing its performance and extensibility.
Functionalities
- Data Import/Export
- Image formats: DICOM, NIFTI, TIFF, JPEG, BMP, PNG
- Support DICOM slice sorting, automatic orientation correction
- DICOM tag editing
- DICOM file manager
- Data Manipulation and Filtering
- Handling 8 bits/16 bits images. And 32bit NIFTI will be converted to 16bits.
- Filters: Sharpness, Gaussian Blur, Enhance Detail, Enhance Focus, Laplacian, Window/Level, Bilateral Filter, Tone Reproduction, Brightness/Contrast, Color Model, Equalization, K-Means Clustering, Gradient, Noise Reduction, Edge Detection, Level Set
- Mask Cropping
- Region Labeling
- Cropping / Resizing / Rotation
- Arbitrary arithmetic operations
- Volume Rendering
- High quality pre-integrated volume rendering by graphics hardware
- Interactive manipulation of transfer function
- Composition of multiple transparent volumes
- Support for stereoscopic viewing
- Viewing & Navigation
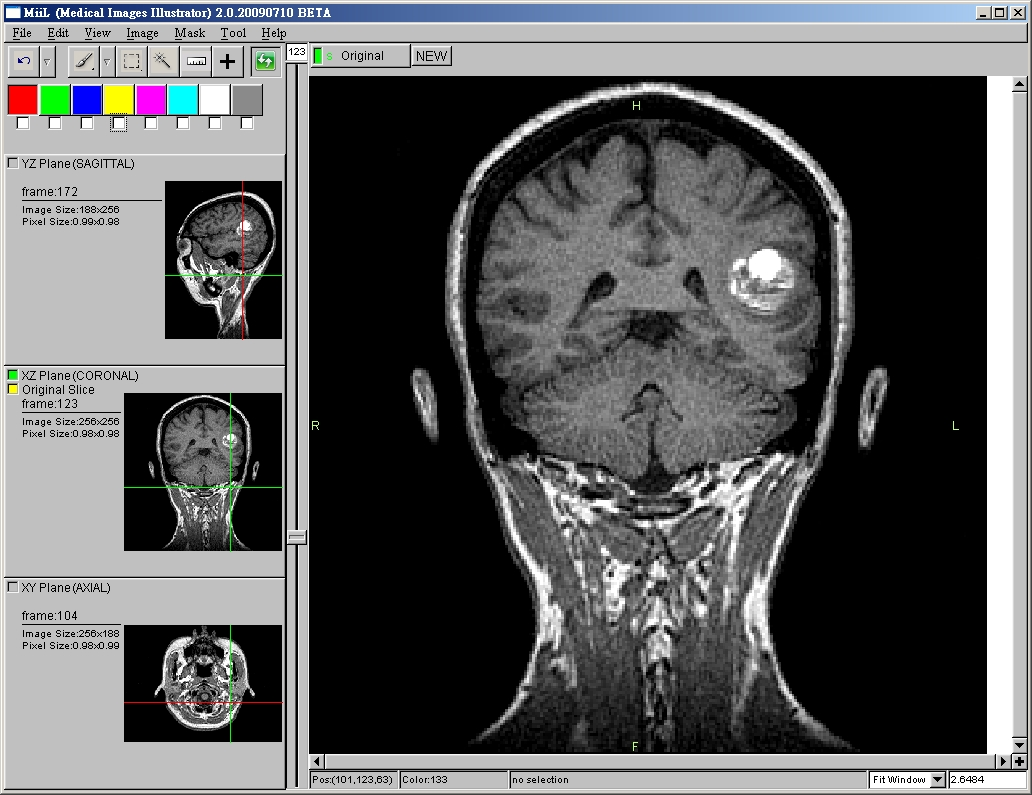
- Slice Viewing with Zooming
- 3 Anatomical Planes (Sagittal, Coronal, Axial) Viewing Synchronization
- Window/Level
- Tiled Window Viewing
- DICOM Tags Viewing
- Data Analysis
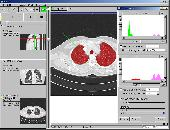
- Histogram analysis
- Probing of data values
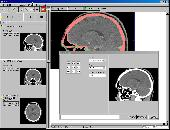
- Measurement of angles, distances, area size, volume size
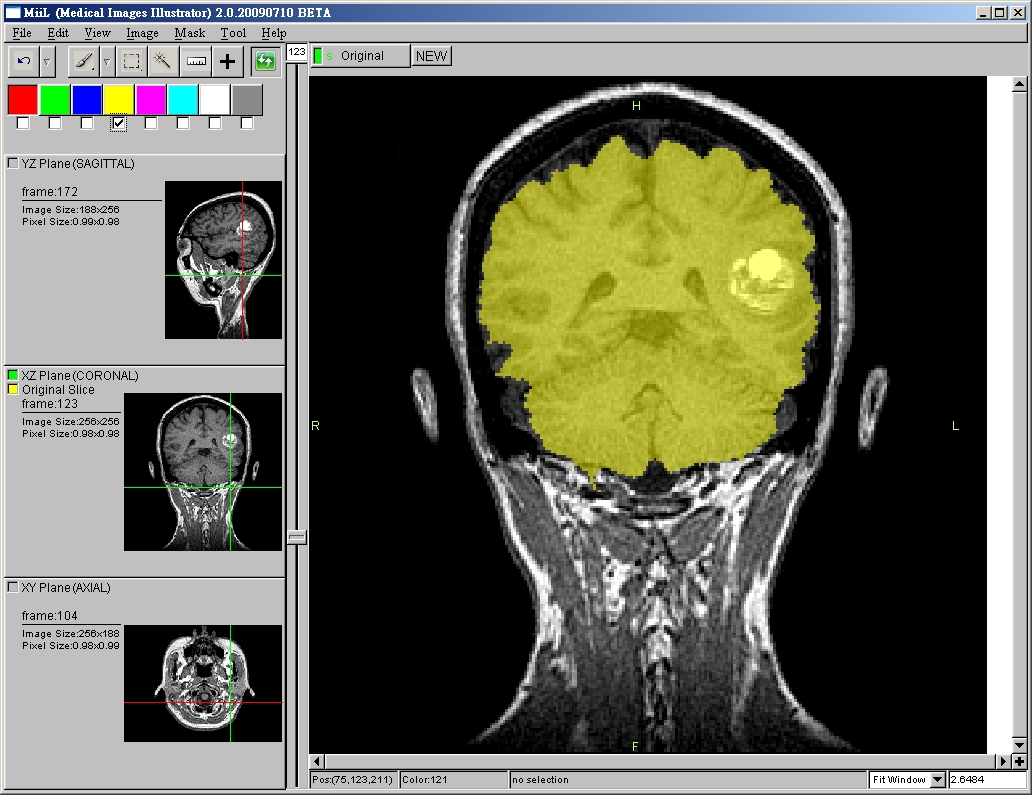
- Image Segmentation
- Segmentation by mask
- Brush tool (painting) with Undo (2D)
- Automatic mask generation by threshold/histogram in 3D
- Magic wand (region growing) in both 2D (slice by slice)
- 3D Seed Growing
- Lazy Snapping
- Watershed
- Mask smoothing, hole filling and island removal in both 2D (slice by slice) and 3D
- Arbitrary arithmetic mask operations in 3D
- Maximum mask region pick up
- Mask mapping between multiple 3D image sets
- Mask loading and saving
- Semi-automatic segmentation for Lung
- DICOM Annotation
- Image Registration
- Landmark based method
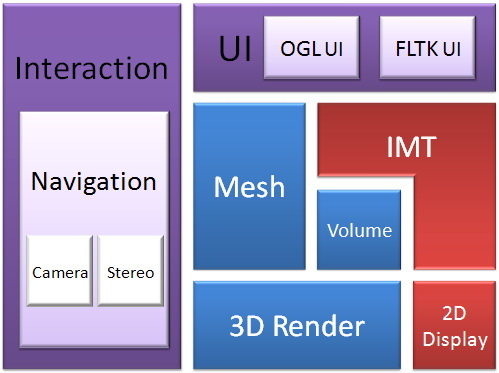
Framework
MiiL is developed based on VIML’s IMT and 3D VR Engine libraries.
Tutorial
Tutorial Video(EN): course page
Tutorial Video(中文): 課程頁面
